Exploring alternate states and oligomerization preferences of coiled-coils by de novo structure modeling
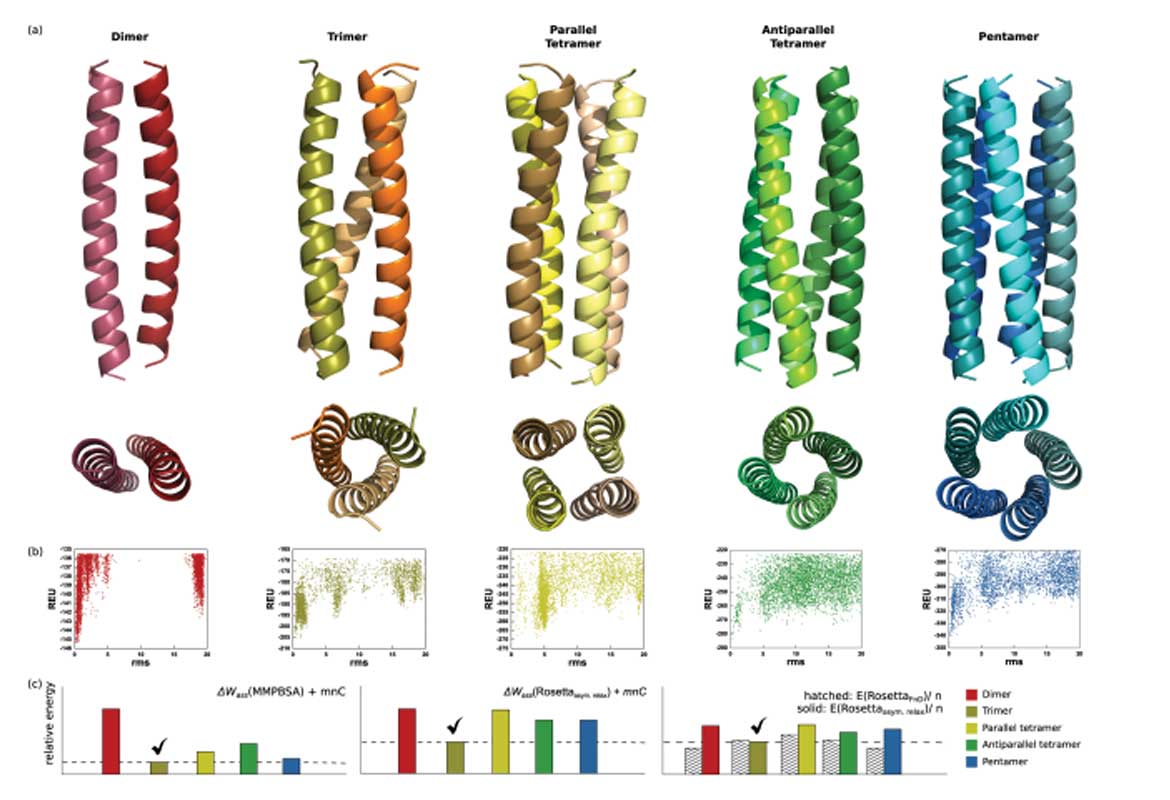
Homomeric coiled-coils can self-assemble into a wide range of structural states with different helix topologies and oligo- meric states. In this study, we have combined de novo structure modeling with stability calculations to simultaneously pre- dict structure and oligomeric states of homomeric coiled-coils. For dimers an asymmetric modeling protocol was developed. Modeling without symmetry constraints showed that backbone asymmetry is important for the formation of parallel dimeric coiled-coils. Collectively, our results demonstrate that high-resolution structure of coiled-coils, as well as parallel and antiparallel orientations of dimers and tetramers, can be accurately predicted from sequence. De novo modeling was also used to generate models of competing oligomeric states, which were used to compare stabilities and thus predict the native stoichiometry from sequence. In a benchmark set of 33 coiled-coil sequences, forming dimers to pentamers, up to 70% of the oligomeric states could be correctly predicted. The calculations demonstrated that the free energy of helix fold- ing could be an important factor for determining stability and oligomeric state of homomeric coiled-coils. The computa- tional methods developed here should be broadly applicable to studies of sequence-structure relationships in coiled-coils and the design of higher order assemblies with improved oligomerization specificity.
