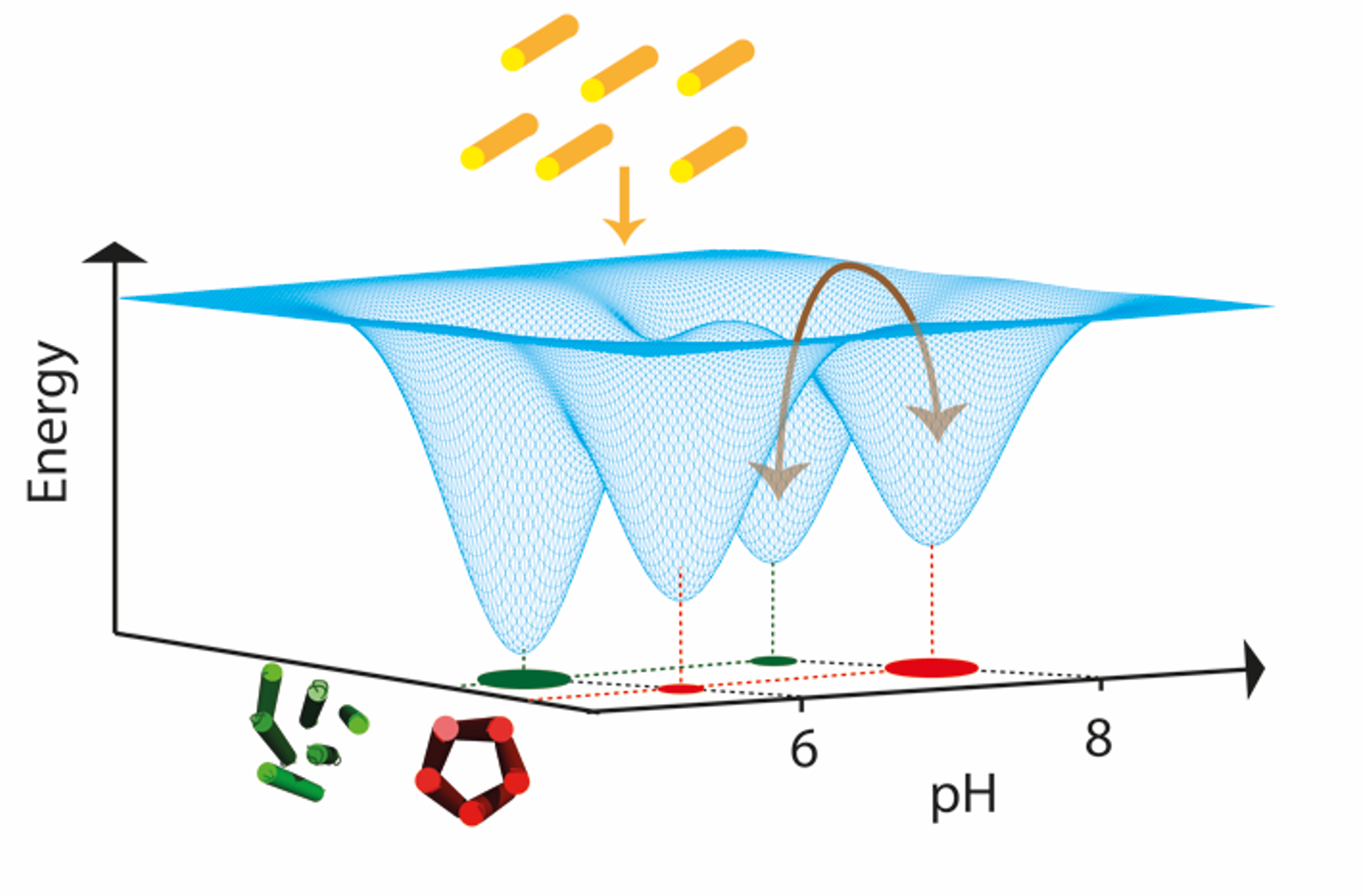
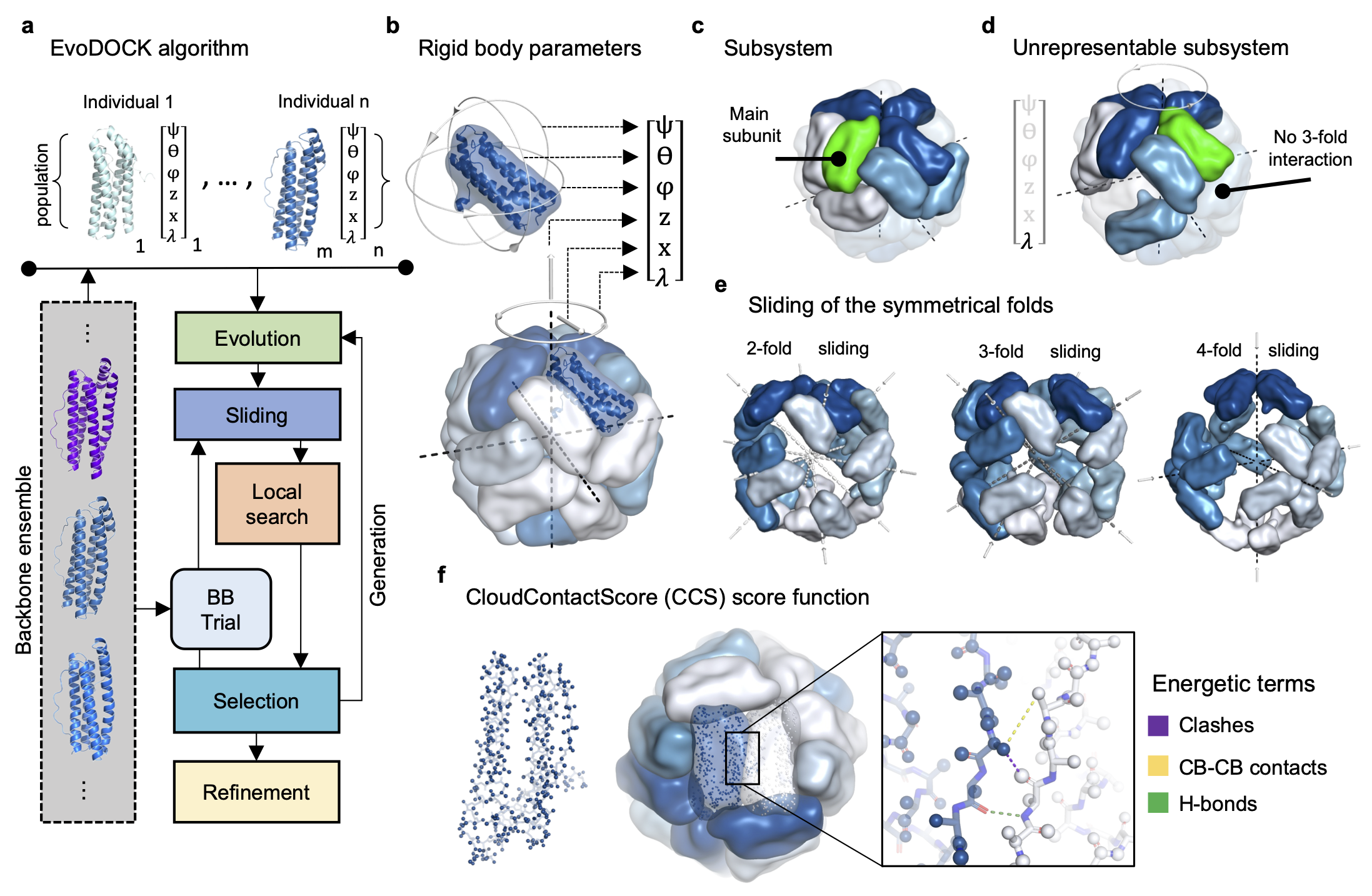
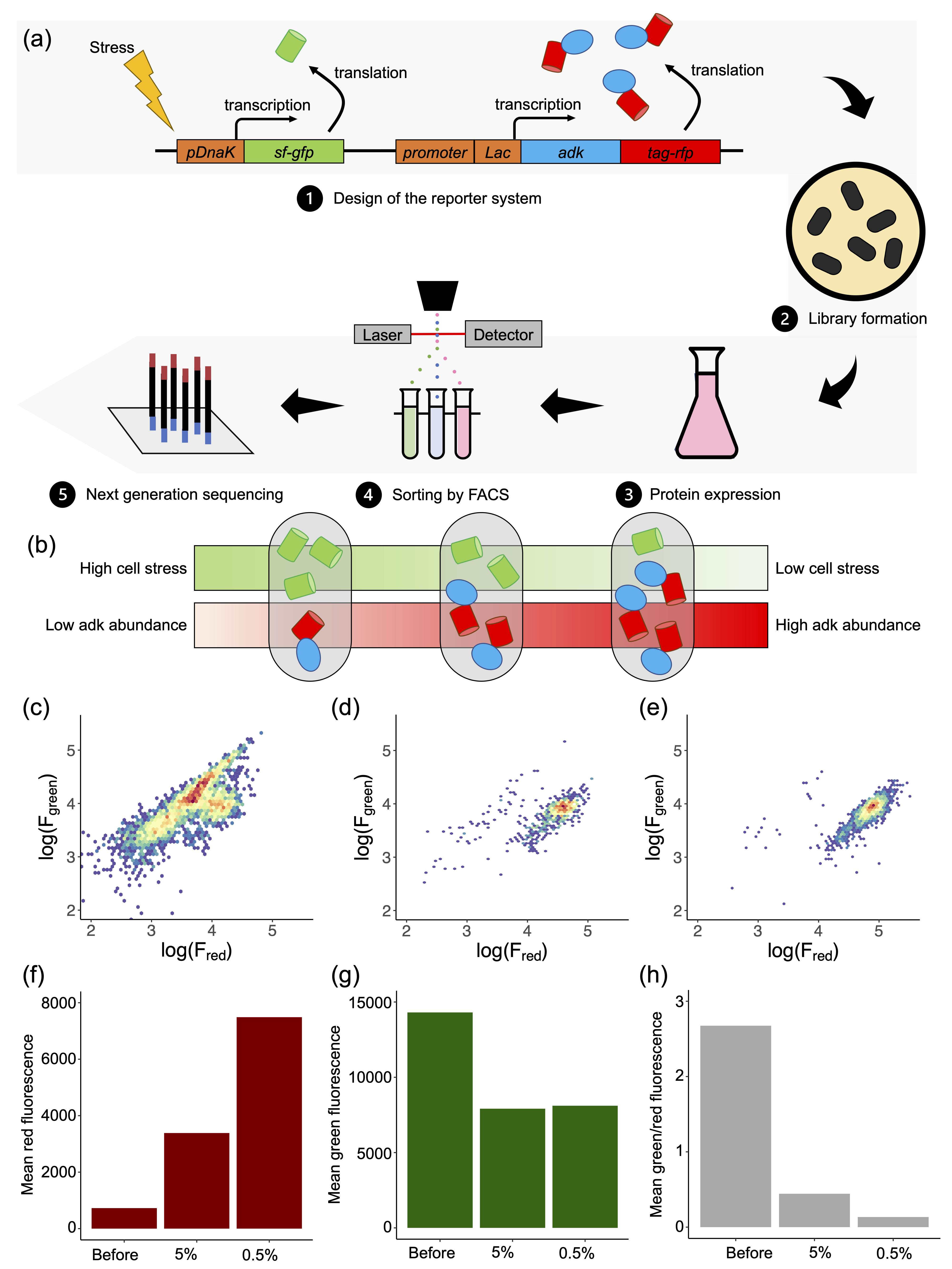
Trajectories in landscapes of sequence, structure and assembly of proteins
Research goals
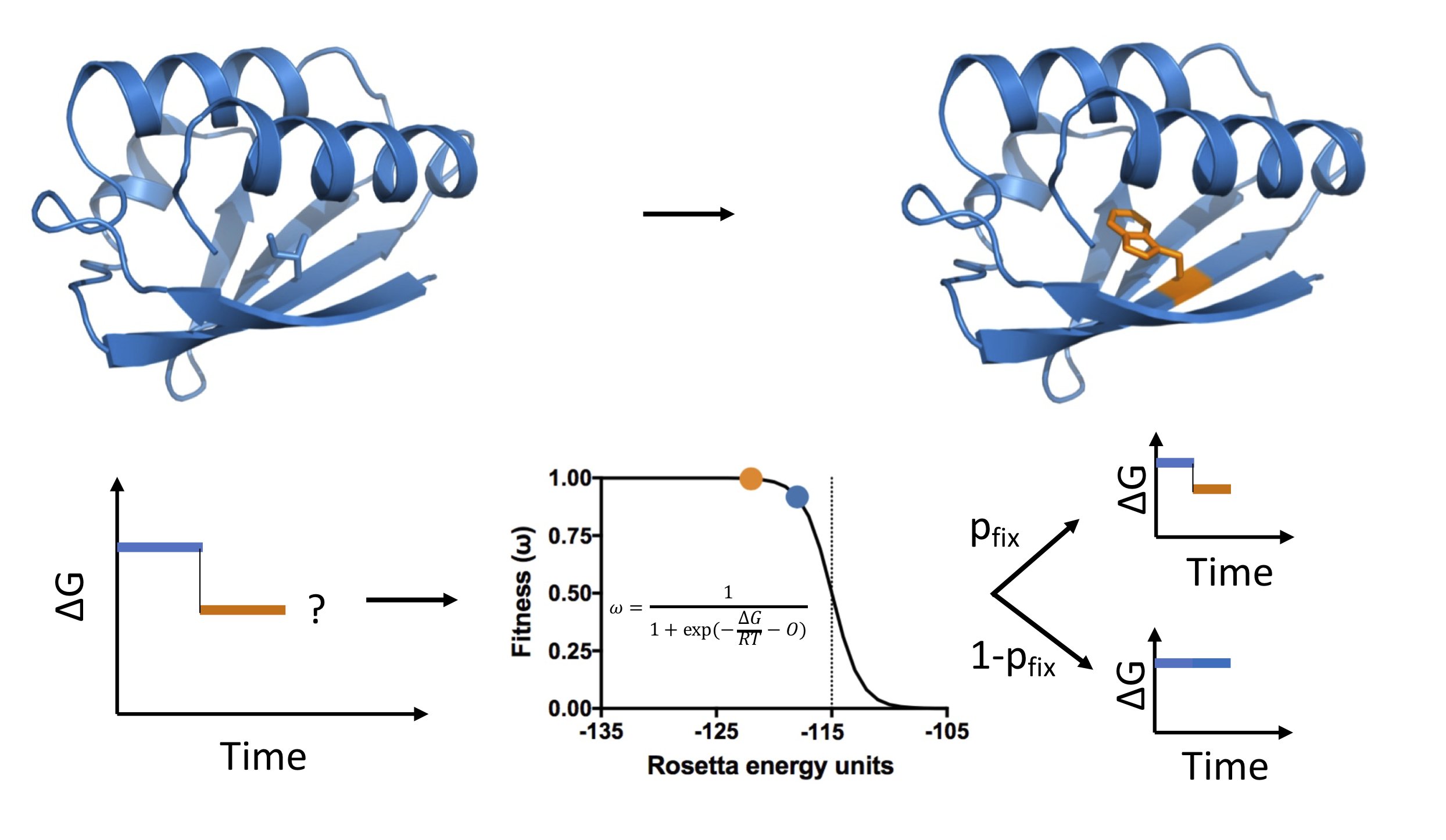
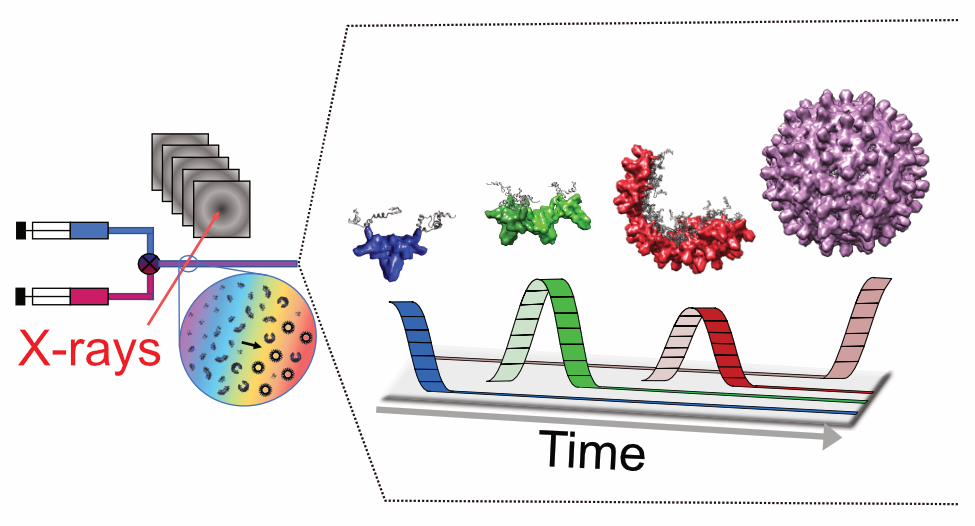
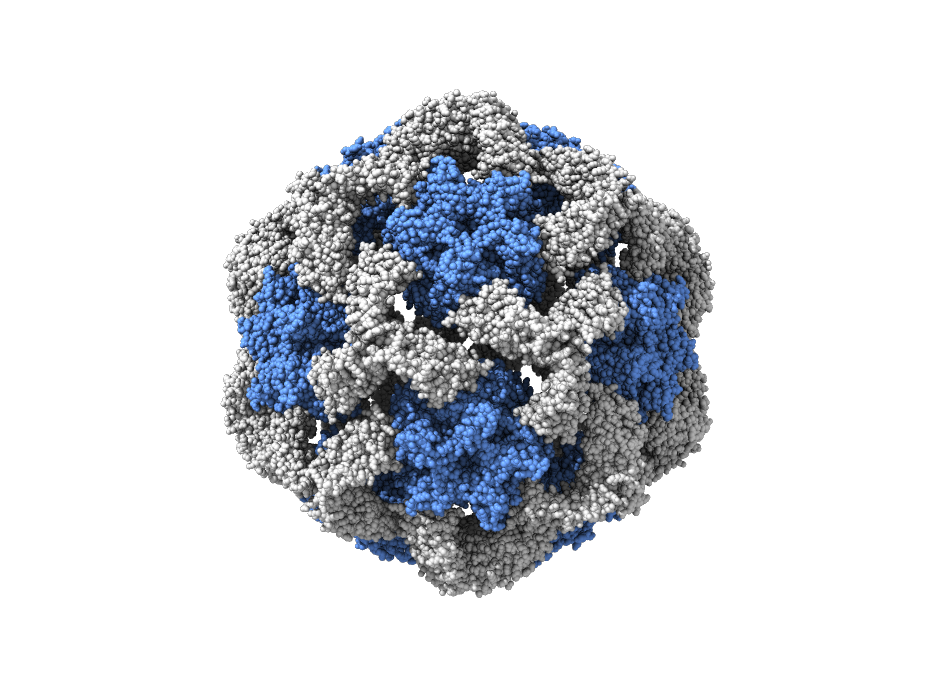
We combine computational and experimental methods to understand the structure, interactions and evolution of proteins. A fundamental goal is to encode our understanding of proteins into computational and experimental methods that enables the design self-assembling protein systems. We also want to understand the process of self-assembly by inferring self-assembly pathways from simulated protein structures and time-resolved experimental data.